RT-LAMP
protocol with open access reagents
Open access means freedom of information. With this protocol, anyone in the world can make their own RT-LAMP assays on a massive scale, producing most of the reagents themselves. Buffers for our open access assays are non-proprietary and their composition and preparation are listed here. Enzymes that power these reactions can be expressed and purified in any molecular biology lab in the world. A plasmid and four days of work is all it takes to produce enough enzymes for hundreds of thousands of RT-LAMP tests for SARS-CoV-2. The open access approach really enables large scale, decentralised and supply chain independent testing strategies. Universities, biohacker collectives, governmental research institutions and many more are encouraged to take matters into their own hands and establish their own, robust production pipelines all over the world.
Here's how it works...
Get plasmids
Order plasmids for enzyme production. Our plasmids are on Addgene for anyone to use.
Express and purify
Follow the published protocols to express enzymes for lots of reactions.
Benchmark and QC
Test your enzyme batches for activity and whole, assembled RT-LAMP reactions for sensitivity.
Test for covid-19
Help secure easy, fast, and cheap testing for your community.
Enzyme expression and purification
RT-LAMP reactions are catalysed by two enzymes, a reverse transcriptase that converts RNA to DNA, and a polymerase, that amplifies DNA in the presence of complementary primers. There are many enzymes that can do this, but non-patented wild-type enzymes best suited for this task are HIV-1 reverse transcriptase (HIV-RT) from the human immunodeficiency virus, and Bst polymerase Large Fragment, from the bacterium Bacillus stearothermophilus. Below, you can find the protocol to express these enzymes in E. coli and how to purify them.
Buffers and solutions
"DANUBE" INACTIVATION SOLUTION
Danube is an inactivation buffer based on the HUDSON (Heating Unextracted Diagnostic Samples to Obliterate Nucleases) protocol. Danube can be prepared as a 10X solution, enabling a larger sample input. Heat inactivation of samples by adding 1 volume of Danube to 9 volumes of sample and heating the mixture to 95 ºC for 5 minutes results in non-infectious samples with a complete inactivation of RNases.
- Danube inactivation buffer is named after the river flowing through Vienna!
- If you don't have these stock solutions on hand, you can prepare them yourself as long as you have analytical scales and other lab equipment. If you are not sure how this is done, google "how to make 0.5 M EDTA solution" for example.
- Use the prepared inactivation solution immediately
Materials
- Tris(2-carboxyethyl)phosphine hydrochloride solution, 0.5 M concentration, pH adjusted to 7.0 with ammonium hydroxide
- Ethylenediaminetetraacetic (EDTA) solution, 0.5 M, pH 8.0
- Betaine, 5 M solution
- Proteinase K, 20 mg/ml, aqueous solution
Procedure
- Calculate the volume of “Danube” inactivation solution you need based on the amount of samples you plan to inactivate.
For performing RT-LAMP, we recommend inactivating at least 10 microlitres of sample, which requires 1 microlitre of Danube per sample. For performing bead-LAMP, we recommend inactivating at least 100 microlitres of sample, which requires 10 microlitres of Danube. - Calculate the volume of needed reagents for number of samples you want to inactivate based on the following table
Reagent Stock concentration 10x concentration Volume to add for 1 ml TCEP 0.5 M 25 mM 50 μl EDTA 0.5 M 10 mM 20 μl Betaine 5 M 4.15 M 830 μl Proteinase K 20 mg/ml 2 mg/ml 100 μl Final Volume 1 ml - Pipette the reagents together and mix by pipetting up and down or by vortexing and centrifuging.
- Use for inactivation of samples immediately.
Isothermal amplification buffer 10X
Isothermal amplification buffer is a well-known reagent in LAMP reactions. It provides optimal reaction conditions for a variety of polymerases derived from BstLF and the wild-type BstLF enzyme as well. A finished, ready to use 10X buffer can be purchased from New England Biolabs, but its composition is publicly known and the buffer itself is very easy to assemble.
- You can order Isothermal Amplification Buffer from NEB or make it yourself from stock solutions in a laboratory.
- If you don't have these stock solutions on hand, you can prepare them yourself as long as you have analytical scales and other lab equipment. If you aren't sure how this is done, Google "how to make 3 M potassium chloride" for example.
- Store the prepared and sterile filtered buffer at -20 ºC.
Materials
- Tris hydrochloride, pH 8.8, 1.5 M solution
- Potassium chloride, 3 M solution
- Ammonium sulfate, 1 M solution
- Magnesium sulfate, 1 M solution
- Tween® 20
- Nuclease-free water
Procedure
- Prepare the materials either as pre-made stock solutions, or by making your own stock solutions.
- Calculate the volume of needed reagents for number of samples you want to inactivate based on the following table
Reagent Stock concentration 10x concentration Volume to add for 1 ml Tris hydrochloride 1.5 M 200 mM 133 μl Potassium chloride 3 M 500 mM 167 μl Ammonium sulfate 1 M 100 mM 100 μl Magnesium sulfate 1 M 20 mM 20 μl Tween® 20 100 % 1 % 10 μl Nuclease-free water 570 μl Final Volume - Pipette the reagents together and mix by pipetting up and down thoroughly or by vortexing and centrifuging.
- Filter through a sterile 0.22 μm syringe filter and store at -20 ºC.
LAMP primer mix 10X
LAMP requires six primers for optimal functioning. That is six short DNA oligonucleotides to synthesise or order, the names are F3, B3, LF, LB, FIP and BIP. To make things easier, one can mix those in a pre-determined ratio to make a 10X primer mix to use with LAMP. You can make this mix with As1 primers against SARS-CoV-2 genome or against human ACTB transcript. Aliquot and label the finished primer mix with the name, date and name of the person who prepared it. We encourage to test every primer mix you receive for activity against the target template, especially when ordering from vendors you don’t have prior experience with.
- Store primer mixes at -20 ºC.
- Order oligonucleotides from reputable companies specialising in DNA oligonucleotide synthesis.
- When ordering oligos, choose "standard desalting" as the purity. We recommend ordering primers resuspended in 100 μM concentration. You can order yours lyophilised and then resuspend them in nuclease-free water according to the supplier's instructions.
- This procedure is applicable for every LAMP primer set we recommend here.
Materials
SARS-COV-2 PRIMER SEQUENCES – As1 primers:
As1_F3 CGGTGGACAAATTGTCAC
As1_B3 CTTCTCTGGATTTAACACACTT
As1_LF TTACAAGCTTAAAGAATGTCTGAACACT
As1_LB TTGAATTTAGGTGAAACATTTGTCACG
As1_FIP TCAGCACACAAAGCCAAAAATTTATCTGTGCAAAGGAAATTAAGGAG
As1_BIP TATTGGTGGAGCTAAACTTAAAGCCCTGTACAATCCCTTTGAGTG
POSITIVE CONTROL PRIMER SEQUENCES – ACTB primers:
ACTB-F3 AGTACCCCATCGAGCACG
ACTB-B3 AGCCTGGATAGCAACGTACA
ACTB-FIP GAGCCACACGCAGCTCATTGTATCACCAACTGGGACGACA
ACTB-BIP CTGAACCCCAAGGCCAACCGGCTGGGGTGTTGAAGGTC
ACTB-LoopF TGTGGTGCCAGATTTTCTCCA
ACTB-LoopB CGAGAAGATGACCCAGATCATGT
procedure
- Prepare your ordered oligonucleotides. If they are not resuspended, resuspend them in nuclease-free water for 100 μM stock concentration.
- In a clean and template-free workspace, such as in a PCR hood, mix the oligonucleotides in the order outlined by the table below. Make sure to add the components from largest volume to the smallest volume.
Oligo name Add for 100 μl of mix F3 2 μl B3 2 μl LF 4 μl LB 4 μl FIP 16 μl BIP 16 μl nuclease-free water 56 μl - Mix the finished primer mix by thoroughly pipetting up and down or by vortexing and briefly spinning down in a microfuge.
- Distribute the finished primer mix into Eppendorf tubes into conveniently sized aliquots, label the tubes and store at -20 ºC.
RT-LAMP protocol
When you have the enzymes, primer mix and the sample inactivation reagent, it’s time to assemble the reactions themselves. Make sure to test your reactions on a known positive sample dilution first to assess the specificity and sensitivity of your assays. Expressing and purifying your own enzymes is not trivial, but the payoff is tremendous. If you are expressing your enzymes and having issues with the resulting reactions, please contact us for help in troubleshooting. Please consider the following before you start:
- SARS-CoV-2 is a highly contagious virus and requires personal protection with appropriate safety standards. Please follow Biosafety level guidelines outlined by the CDC or your local authorities.
- RT-LAMP reactions are incompatible with samples in collection media containing more than 1 mM of metal chelators such as EDTA or EGTA and with sample collection media containing chaotropic reagents such as guanidium thiocyanate and guanidium hydrochloride.
- Separate working areas for sample preparation, reaction set-up, and analysis are highly recommended. Cleaning work surfaces and pipettes with 5-10% bleach solution.
- Do not open RT-LAMP reactions in order to prevent carry-over cross contamination leading to false positives. The addition of dUTP and Antarctic Thermolabile UDG enzyme to the reaction greatly reduces the contamination risk, therefore we strongly recommend adding them to your RT-LAMP reactions.
- 96-well plates must be properly sealed. This is absolutely crucial to prevent cross-well contamination. Proper plate seals (e.g. tight sealing plastic covers or aluminium seals), a proper plate-roller (e.g. BioRad) or even better a PCR plate sealer (e.g. BioRad PX1 PCR plate sealer) can help prevent cross-well contamination.
- Store reaction components at the appropriate temperatures outlined in the protocol or by the manufacturer. It is crucial to thoroughly mix all components prior to use to ensure the homogeneity of the reagents.
- Colorimetric and fluorescent detection can be performed in the same reaction by simple addition of both dyes (the colorimetric HNB dye and SYTO-9 fluorescent dye that is listed in the protocol as optional).
- When working with crude samples lysed in Danube inactivation buffer, keep samples on ice whenever possible. While we have observed complete inactivation of nucleases after 5 minutes at 95°C, we found considerable RNase activity in biological sample types such as saliva and gargle. Due to the FBS content in Universal Transport Media (UTM), nasopharyngeal swabs in this medium may also contain RNases.
- RT-LAMP uses six primers, two inner loop primers (FIP, BIP), loop primers (LF, LB) and outer bumping primers (F3, B3). A reaction contains all six primers if a single amplicon is tested.
- A positive control amplicon detecting the human ACTB gene can be used to confirm the presence of human nucleic acid sample material in the sample, and the absence of general reaction-inhibiting substances in the sample.
- A crucial component of every experiment and testing procedure is the selection of proper controls. For verifying that your reaction mastermix works in the given conditions for the detection of SARS-CoV-2 genome, we recommend the use of inactivated qPCR positive patient sample, synthetic SARS-CoV-2 RNA or plasmid DNA containing the Orf1ab region. Positive controls should have a concentration exceeding 100 copies/µl. Verified qPCR negative sample is a proper choice for a negative control.
- The enzymes you produce are likely different in activity from those that other labs produce. Make sure to test your enzymes for activity.
- The concentration of your enzymes will differ from the one used in this protocol. Take this into account and make sure to add enough to match the final concentrations in the table below.
- Dilute enzymes in the dilution buffer as described in the enzyme expression and purification protocol to preserve their activity and protect them from degradation.
- Store enzymes at -80ºC in single use aliquots and work with enzymes on ice.
SAMPLE INACTIVATION AND LYSIS USING DANUBE INACTIVATION SOLUTION (BSL2 WORKSPACE)
- Prepare “Danube” 10X inactivation solution according to the protocol above.
- In a BSL-2 workspace with extra precautions taken against respiratory viruses, mix 10μl of Danube inactivation solution with 90 μl of sample.
[CRITICAL] Make sure to adhere to the safety precautions for handling potentially infectious SARS-CoV-2 samples outlined by applicable health authorities in your jurisdiction. - Heat-inactivate the mixture of sample and inactivation solution at 95°C for 5 minutes.
- Cool the inactivated samples on ice or at room temperature for immediate use or store at -20°C.
[PAUSE] Samples can be stored at -20°C at this point for up to four weeks, potentially longer. Inactivated samples can now be handled in BSL-1 settings.
RT-LAMP reaction assembly (BSL1 WORKSPACE)
- Calculate your reagent consumption for assembling the RT-LAMP reaction master mix according to the table below. Multiply the last column by the number of samples you plan to test + 2 for a positive and negative control. Multiply this number by 1.1 to make sure you prepare an excess to compensate for pipetting error.
Reagent Stock concentration Final concentration To add per reaction Isothermal amplification buffer 10X 1X 1 μl dNTP mix 25 mM 1.4 mM 0.56 μl dUTP 100 mM 0.7 mM 0.07 μl Magnesium sulfate 100 mM 6 mM 0.6 μl As1 primer mix 10X 1X 1 μl HNB dye, trisodium salt 20 mM 0.12 mM 0.06 μl Syto9 fluorescent dye (optional) 100 μM 2 μM 0.2 μl Betaine 5 M 0.4 M 0.8 μl HIV-RT enzyme 0.55 mg/ml 0.00275 mg/ml 0.05 μl BstLF enzyme 0.6 mg/ml 0.02 mg/ml 0. 33 μl Thermolabile UDG enzyme 1 U/μl 0.02 U/μl 0.2 Nuclease-free water to a total of 8 μl Final Volume 8 μl - Prepare the RT-LAMP reaction master mix by mixing the reagents in an Eppendorf tube on ice. Add the enzymes last.
[CRITICAL] Make sure to add the enzyme components last! They will otherwise denature and you will experience decreased assay performance or no activity at all.
[CRITICAL] Assemble the RT-LAMP reaction master mix on ice. - Mix the reagents by thoroughly pipetting up and down or by vortexing and spin down in a microfuge.
- Dispense the reaction mix into PCR strips or into a 96-well plate on ice. Each well should receive 8 μl of the reaction mix.
- On ice, add 2 μl of heat-inactivated sample treated with Danube buffer to RT-LAMP reaction mix and mix the reaction by pipetting up and down 10 times. Repeat for all samples that are to be tested, and include a proper positive and negative control. Seal PCR strips by capping them, and seal 96-well plates by applying a plastic transparent plate seal.
[CRITICAL] This step should be done in a separate work area where no reagents are handled.
[CRITICAL] If performing colorimetric detection, take an image of the PCR strips or 96-well plate with the mastermix and samples. - Transfer the reactions to a suitable stable heat-source such as a heat block, thermocycler or a water bath. Incubate the reactions at 63°C for 35 minutes.
[CRITICAL] For highest sensitivity and specificity, make sure you transfer the sample from ice to a preheated, 63°C environment.
[OPTIONAL] For real-time fluorescence data acquisition in a qPCR thermocycler, we typically perform 35 cycles at 63°C with 1-minute cycle length and reading at the end of each cycle. Syto9 fluorescent dye requires data acquisition using a standard FAM or SYBR filter (494nm/518nm absorption/emission). - After 35 minutes run-time, remove reactions and allow to cool briefly at room temperature. Then proceed to inspect reactions visually. Negative reactions are purple while positive reactions are sky-blue.
- Discard the reactions immediately after noting down the results.
[CRITICAL] If performing colorimetric detection, take a second image of the PCR strips or 96-well plate under the same conditions as you took the first one after the reaction. - [CRITICAL] DO NOT OPEN THE REACTION TUBES!
Readout methods
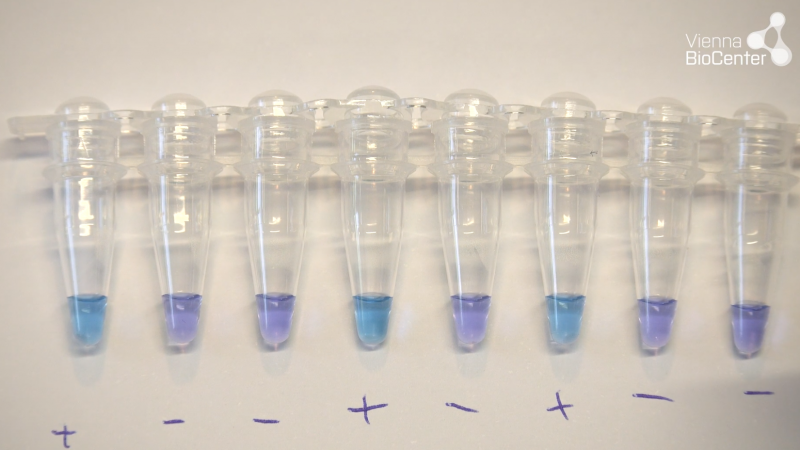
Visual colorimetric
Hydroxynaphtol blue (HNB) is used in RT-LAMP reactions that do not include any purification step. The dye changes colour from purple to sky blue upon presence of target sequence and amplification. This can be visualised with the naked eye, and reactions can be scored directly after taking them out of the heating device.

App-assisted colorimetric
For people who find the colour change of HNB difficult to see with the naked eye, we recommend the use of a simple web app. Developed by Andrew Straw to increase the colour difference between positives and negatives and make the readout much simpler. Try it out on colorimetry.net

Fluorimetric
Amplification of DNA in RT-LAMP reactions can also be read out through the use of fluorescent intercalating dyes. These dyes, such as Syto9 or the NEB LAMP dye, enable the user to either monitor the progress of the reaction in real time through the use of a qPCR machine, or end-point detection with a plate reader or a similar fluorescence detection device.
Premixing information
Assembling each master mix for every testing session from scratch is not necessary, however freezing prepared master mixes increases the risk of running into sensitivity or specificity issues. We’ve tested the possible combinations and now we can tell you what you can and can’t premix for easier reaction assembly without compromising on the quality of the tests.
LAMP reaction buffer premix
| Reagent | Stock concentration | To add per 100 reactions (10µl size) | |
|---|---|---|---|
| Isothermal amplification buffer | 10X | 100 μl | |
| dNTP mix | 25 mM | 56 μl | |
| dUTP | 100 mM | 7 μl | |
| Magnesium sulfate | 100 mM | 60 μl | |
| As1 primer mix | 10X | 100 μl | |
| HNB dye | 20 mM | 6 μl | |
| Syto9 fluorescent dye (optional) | 100 μM | 20 μl | |
| Betaine | 5 M | 80 μl | |
| nuclease-free water | 321 μl | ||
| total | 750 μl |
Add 7.5 µl of this mix per 1 reaction.
Store at -20 °C.
LAMP enzyme premix
| Reagent | Stock concentration | Amount per reaction | To add per 100 reactions (10µl size) |
|---|---|---|---|
| HIV-RT enzyme | variable | 75 ng | variable |
| BstLF enzyme | variable | 200 ng | variable |
| Thermolabile UDG enzyme (NEB) | 1 U/μl | 0.2 U | 20 μl |
| storage buffer | to a total of 50 µl | ||
| total | 50 µl |
Add 0.5 µl of this mix per 1 reaction.
The volumes for enzymes are listed as “variable” since each enzyme purification yields a different concentration of enzyme. We recommend measuring the yield you get from your purifications and make the calculations yourself, so as to reach 75 ng of reverse transcriptase and 200 ng of DNA polymerase, respectively. Then multiply the volumes for 75 ng and 200 ng of enzyme, respectively, by 100 to fill in the table. Add storage, as described in the enzyme expression and purification protocol on the open access protocol pages, to a total of 50 µl.
Store at -20 °C.
The premixes for open-access RT-LAMP enable you to have two tubes in your -20 °C freezer and to assemble reactions easily using 7.5 µl of LAMP reaction buffer premix and 0.5 µl of LAMP enzyme premix per 10 µl reaction. We’ve observed their stability to be unchanged after 1 week’s storage at -20 °C, but they can most likely last for longer. It is crucial to work quickly with the enzyme mix and to work with it on ice or in the cold.
